- Objective
- Project Tools
- Project steps
- Application link
- Sources
- Acknowledgments
Objective
Using The Global Health Observatory OData API to fetch data from it’s data collection. Analyze the data using data science techniques and create an interactive dashboard and deploying it using cloud services.
Project Tools
Data Source:
- The Global Health Observatory https://www.who.int/data/gho
- OData API documentation: https://www.who.int/data/gho/info/gho-odata-api
Retrieve the data:
- requests to make HTTP requests to the OData API and retrieve the data.
- Pandas: Perform data preprocessing, cleaning, filtering, and transformation using the powerful data manipulation capabilities of pandas.
Analyze the Data:
- Pandas: Utilize pandas for exploratory data analysis (EDA), data wrangling, and statistical analysis on the retrieved data.
- Stats: Module that contains correlation functions and statistical tests among other fuctions.
Visualization Tools:
- Matplotlib: Create static visualizations such as line plots, bar charts, and scatter plots.
- Seaborn: Build aesthetically pleasing and informative statistical visualizations.
- Plotly: Develop interactive and customizable visualizations, including interactive charts and dashboards.
Design the Dashboard:
- Dash: Useful to build interactive web-based dashboards using Python and HTML components.
Deploy the Dashboard:
- AWS cloud services
- TMUX library.
Test and Iterate:
- User feedback and testing: Gather feedback from potential users and testers to improve the functionality and user experience of your dashboard.
- Jupyter Notebook or documentation: Document the project, including the data analysis process, code, and key findings.
Project steps
-
Select the Data Source: Determine the specific API that provides the data needed for the analysis. Explore available public datasets. Determine the analysis goals.
-
Understand the API: Study the documentation of the OData API to understand its endpoints, query parameters, and how to retrieve data. Identify the available resources, data models, and relationships between entities.
-
Retrieve and Preprocess Data: Use the OData API to fetch the required data based on the analysis goals. Apply data preprocessing steps like cleaning, filtering, and transforming the data as necessary to prepare it for analysis.
-
Analyze the Data: Utilize data science techniques and libraries such as pandas, stats to perform exploratory data analysis (EDA), statistical analysis and visualizations. Extract meaningful insights and identify patterns or correlations in the data. Select the appropriate visualization tool like Matplotlib, Seaborn, or Plotly to create interactive and informative visualizations.
-
Design the application/dashboard: Design the user interface and layout of the dashboard using Dash library. Create an intuitive and user-friendly dashboard that allows users to interact with the visualizations, apply filters, and explore the data dynamically. Iterate on the design and make improvements based on user input and additional data analysis requirements.
-
Deploy the Dashboard: Host the dashboard on a web server or a cloud platform to make it accessible online. This involves deploying it as a web application utilizing AWS cloud hosting services. Test and Iterate: Test the functionality and usability of the dashboard, seeking feedback from potential users.
Select the Data Source
What I found exploring the GHO Data Collection:
- The GHO data repository serves as the World Health Organization’s (WHO) portal to health-related statistics concerning its 194 Member States.
- The Data could be explored by Theme, Category or Indicator.
- The GHO data repository offers access to a wide range of health indicators, exceeding 2000 in number.
- Indicators cover crucial health topics such as mortality, disease burden, Millennium Development Goals, non-communicable diseases, infectious diseases, health systems, environmental health, violence, injuries, and equity, among others.
- Several indicators do not contain data or the data is not updated or completed.
Understanding the API
The Global Health Observatory (GHO) is constructed using a technology called RESTful web service. REST stands for Representational State Transfer, which is an architectural style used to design networked applications. In this context, GHO uses RESTful web services as the underlying technology to provide access to its data.
RESTful web services use standard HTTP methods (such as GET, POST, PUT, DELETE) to interact with resources (data) over the internet. These resources are identified by URLs, and the web service responds with data in a structured format, often in JSON or XML.
By using a RESTful web service, the Global Health Observatory can allow users to easily retrieve health-related data through standard web protocols, making it accessible and interoperable across various platforms and programming languages. This approach enables efficient data access and manipulation, making it convenient for data analysts and developers to fetch and utilize the health data provided by the GHO.
- The documentation is clear and extensive.
Retrieve and Preprocess Data:
Use the OData API to fetch the required data based on the analysis goals. Apply data preprocessing steps like cleaning, filtering, and transforming the data as necessary to prepare it for analysis.
Retrieving
# Import the libraries
import pandas as pd
import requests
import json
import plotly.express as px
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
GET request to obtain the dimension data directly from the Global Health Observatory API
Retrieving all Dimension Values for Country dimension
# Send a GET request to the API endpoint
url = "https://ghoapi.azureedge.net/api/DIMENSION/COUNTRY/DimensionValues"
response = requests.get(url)
# Check the response status code
if response.status_code == 200:
resp = response.json()
data_countries = resp['value']
# Process the data as needed
for item in data_countries:
value = item
print('Request Successful')
# Process each item in the data
else:
print("Request failed with status code:", response.status_code)
Request Successful
# Create a countries DataFrame
countries = pd.DataFrame(data_countries)
countries.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 248 entries, 0 to 247
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Code 248 non-null object
1 Title 248 non-null object
2 Dimension 248 non-null object
3 ParentDimension 242 non-null object
4 ParentCode 242 non-null object
5 ParentTitle 242 non-null object
dtypes: object(6)
memory usage: 11.8+ KB
countries.head(5)
| Code | Title | Dimension | ParentDimension | ParentCode | ParentTitle | |
|---|---|---|---|---|---|---|
| 0 | ABW | Aruba | COUNTRY | REGION | AMR | Americas |
| 1 | AFG | Afghanistan | COUNTRY | REGION | EMR | Eastern Mediterranean |
| 2 | AGO | Angola | COUNTRY | REGION | AFR | Africa |
| 3 | AIA | Anguilla | COUNTRY | REGION | AMR | Americas |
| 4 | ALB | Albania | COUNTRY | REGION | EUR | Europe |
Retrieving all Indicators (list of indicators)
# Send a GET request to the API endpoint
url = "https://ghoapi.azureedge.net/api/Indicator"
response = requests.get(url)
# Check the response status code
if response.status_code == 200:
resp = response.json()
data_indicators = resp['value']
# Process the data as needed
for item in data_indicators:
value = item
value #print to see values
# Process each item in the data
else:
print("Request failed with status code:", response.status_code)
# Create a indicators DataFrame
indicators = pd.DataFrame(data_indicators)
indicators.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 2369 entries, 0 to 2368
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 IndicatorCode 2369 non-null object
1 IndicatorName 2369 non-null object
2 Language 2369 non-null object
dtypes: object(3)
memory usage: 55.6+ KB
pd.set_option('display.max_colwidth', None)
indicators.sample(10)
| IndicatorCode | IndicatorName | Language | |
|---|---|---|---|
| 1295 | SA_0000001512 | Advertising restrictions at cinemas | EN |
| 735 | NCD_CCS_palliative_home | General availability of palliative care in community or home-based care in the public health system | EN |
| 1333 | SA_0000001534 | On-premise sales restrictions on outlet density | EN |
| 495 | GOE_Q178 | Individuals have the legal right to specify which health-related data from their EHR can be shared with health professionals of their choice | EN |
| 1709 | TOTENV_1 | Deaths attributable to the environment | EN |
| 638 | MDG_0000000015 | Prevalence of condom use by adults during higher-risk sex (15-49) (%) | EN |
| 967 | RADON_Q702 | Mandatory mitigation measures if legal value is exceeded (existing buildings) | EN |
| 1474 | SA_0000001739_ARCHIVED | Alcohol, heavy episodic drinking (15+) past 30 days (%), age-standardized with 95%CI | EN |
| 871 | OCC_8 | Occupational airborne particulates attributable DALYs per 100'000 capita | EN |
| 470 | GOE_Q138 | Lack of suitable courses available | EN |
The number of indicators is really high. Previous exploration of the data (directly in the website) showed that a lot of those indicators do not contain updated data or have several NaN values.
The API is working accordingly and I am pulling the data. Nevertheless the GHO website is not totally coherent with the list of indicators I am working with through the API. This add complexity to the process, considering it is relevant to find the indicator description. Additional description about it could be found in the extended version of this project.
Retrieving Tobacco indicators
Tobacco use is indeed a significant contributor to a wide range of non-communicable diseases (NCDs) and is a leading cause of preventable illness and death worldwide. Non-communicable diseases are chronic conditions that are not caused by infectious agents but rather result from a combination of genetic, lifestyle, and environmental factors. These diseases include cardiovascular diseases (like heart disease and stroke), cancer, chronic respiratory diseases (such as chronic obstructive pulmonary disease), and diabetes. Division of Global Health Protection, Global Health, Centers for Disease Control and Prevention.
The harmful effects of tobacco use are primarily attributed to the various toxic substances present in tobacco products, such as nicotine, tar, and carbon monoxide. These substances can damage organs and tissues in the body and increase the risk of developing NCDs. It’s also worth noting that secondhand smoke exposure, which occurs when non-smokers breathe in smoke from tobacco products, is also harmful and can lead to similar health risks.
According with Canadian Cancer Society Second-hand smoke is what smokers breathe out and into the air. It’s also the smoke that comes from a burning cigarette, cigar or pipe. Second-hand smoke has the same chemicals in it as the tobacco smoke breathed in by a smoker. Hundreds of the chemicals in second-hand smoke are toxic. More than 70 of them have been shown to cause cancer in human studies or lab tests. Every year, about 800 Canadian non-smokers die from second-hand smoke.
# Looking for the indicators related with 'Tobacco'
Tobacco_indicators = indicators[indicators['IndicatorName'].str.contains('tobacco', case=False)]
Tobacco_indicators.sample(5)
| IndicatorCode | IndicatorName | Language | |
|---|---|---|---|
| 979 | Rev_imp_other | Annual tax revenues - import duties and all other taxes (excluding corporate taxes on tobacco companies) | EN |
| 2281 | R_admin_tax_stamps_other | Tax stamps, fiscal marks, banderole or other types of marking are applied on other tobacco products | EN |
| 1769 | W14_harm_effects_C | Do health warnings on smokeless tobacco packaging describe the harmful effects of tobacco use on health? | EN |
| 112 | E12_compliance | Compliance of ban on brand name of non-tobacco products used for tobacco product | EN |
| 1931 | R_earmarking | Reported use of earmarked tobacco taxes | EN |
The indicators related with Tobacco are divided by groups according with the strategy MPOWER. “These measures are intended to assist in the country-level implementation of effective interventions to reduce the demand for tobacco, contained in the WHO FCTC.” Read more clicking here
Tobacco_indicators.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 137 entries, 47 to 2354
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 IndicatorCode 137 non-null object
1 IndicatorName 137 non-null object
2 Language 137 non-null object
dtypes: object(3)
memory usage: 4.3+ KB
There are 137 indicators related with ‘Tobacco’, I’ll choose some on them to explore the data available
# Look for the MPOWER group indicators
MPOWER_indicators = Tobacco_indicators[Tobacco_indicators['IndicatorCode'].str.contains(r'^[A-Za-z]_Group', case=False)]
MPOWER_indicators
| IndicatorCode | IndicatorName | Language | |
|---|---|---|---|
| 104 | E_Group | Enforcing bans on tobacco advertising, promotion and sponsorship | EN |
| 615 | M_Group | Monitoring tobacco use and prevention policies | EN |
| 837 | O_Group | Offering help to quit tobacco use | EN |
| 881 | P_Group | Protecting people from tobacco smoke | EN |
| 935 | R_Group | Raising taxes on tobacco | EN |
| 1751 | W_Group | Warning about the dangers of tobacco | EN |
# Let's explore indicator related with prevalence
Tobacco_prevalence = Tobacco_indicators[Tobacco_indicators['IndicatorName'].str.contains('prevalence', case=False)]
Tobacco_prevalence
| IndicatorCode | IndicatorName | Language | |
|---|---|---|---|
| 614 | M_Est_tob_curr | Estimate of current tobacco use prevalence (%) | EN |
| 1942 | Adult_curr_smokeless | Prevalence of current smokeless tobacco use among adults (%) | EN |
| 2001 | Yth_daily_smokeless | Prevalence of daily smokeless tobacco use among adolescents (%) | EN |
| 2070 | M_Est_smk_curr | Estimate of current tobacco smoking prevalence (%) | EN |
| 2076 | Adult_daily_smokeless | Prevalence of daily smokeless tobacco use among adults (%) | EN |
| 2123 | Yth_daily_tob_smoking | Prevalence of daily tobacco smoking among adolescents (%) | EN |
| 2160 | Adult_curr_tob_smoking | Prevalence of current tobacco smoking among adults (%) | EN |
| 2163 | Yth_curr_smokeless | Prevalence of current smokeless tobacco use among adolescents (%) | EN |
| 2180 | Adult_curr_tob_use | Prevalence of current tobacco use among adults (%) | EN |
| 2256 | Yth_curr_tob_use | Prevalence of current tobacco use among adolescents (%) | EN |
| 2277 | Yth_curr_tob_smoking | Prevalence of current tobacco smoking among adolescents (%) | EN |
| 2311 | M_Est_tob_curr_std | Estimate of current tobacco use prevalence (%) (age-standardized rate) | EN |
| 2324 | Adult_daily_tob_smoking | Prevalence of daily tobacco smoking among adults (%) | EN |
| 2349 | Adult_daily_tob_use | Prevalence of daily tobacco use among adults (%) | EN |
| 2354 | M_Est_smk_curr_std | Estimate of current tobacco smoking prevalence (%) (age-standardized rate) | EN |
There are 15 indicators related with tobacco and prevalence.
I used a for loop to write the urls and explore the data in all the indicators within the word ‘Prevalence’, the process is available in the extended version on GitHub. Finally, the indicator I found more interesting by the amount of data it provides was M_Est_smk_curr_std == Estimate of current tobacco use prevalence (%)
Summarizing the indicators exploration:
Indicators to work with in the dashboard:
| Indicator Code | Indicator Name |
|---|---|
| M_Est_smk_curr_std: | Estimate of current tobacco use prevalence (%) |
| M_Group: | Monitoring tobacco use and prevention policies |
| P_Group: | Protecting people from tobacco smoke |
| O_Group: | Offering help to quit tobacco use |
| W_Group: | Warning about the dangers of tobacco |
| E_Group: | Enforcing bans on tobacco advertising, promotion and sponsorship |
| R_Group: | Raising taxes on tobacco |
| NCD_UNDER70: | Premature deaths due to noncommunicable diseases (NCD) as a proportion of all NCD deaths |
Indicators Dataframes
# Create a for loop to create the urls for the indicators I found interesting.
IndicatorCodes = ['M_Est_smk_curr_std', 'M_Group', 'P_Group', 'O_Group', 'W_Group', 'E_Group', 'R_Group', 'NCD_UNDER70']
urls = []
for Codes in IndicatorCodes:
# Create the URLs
url = f"https://ghoapi.azureedge.net/api/{Codes}"
urls.append(url)
urls
['https://ghoapi.azureedge.net/api/M_Est_smk_curr_std',
'https://ghoapi.azureedge.net/api/M_Group',
'https://ghoapi.azureedge.net/api/P_Group',
'https://ghoapi.azureedge.net/api/O_Group',
'https://ghoapi.azureedge.net/api/W_Group',
'https://ghoapi.azureedge.net/api/E_Group',
'https://ghoapi.azureedge.net/api/R_Group',
'https://ghoapi.azureedge.net/api/NCD_UNDER70']
# Create a for loop to retrieve the data and transform it into dataframes
df_dictionary = {}
for url in urls:
response = requests.get(url)
# Check the response status code
if response.status_code == 200:
resp = response.json()
data = resp['value']
# Transform the json responses into dataframes.
df = pd.DataFrame(data)
# Name the df with the indicator code
df_name = f"df_{url.split('/')[-1]}"
# Include each name df into a dictionary
df_dictionary[df_name] = df
else:
print("Request failed with status code:", response.status_code)
# print the dictionary of dataframes.
print(df_dictionary.keys())
dict_keys(['df_M_Est_smk_curr_std', 'df_M_Group', 'df_P_Group', 'df_O_Group', 'df_W_Group', 'df_E_Group', 'df_R_Group', 'df_NCD_UNDER70'])
# Extract the dfs from the dictionary
df_M_Est_smk_curr_std = df_dictionary['df_M_Est_smk_curr_std']
df_M_Group = df_dictionary['df_M_Group']
df_P_Group = df_dictionary['df_P_Group']
df_O_Group = df_dictionary['df_O_Group']
df_W_Group = df_dictionary['df_W_Group']
df_E_Group = df_dictionary['df_E_Group']
df_R_Group = df_dictionary['df_R_Group']
df_NCD_UNDER70 = df_dictionary['df_NCD_UNDER70']
# print the shape
for df in [df_M_Est_smk_curr_std, df_M_Group, df_P_Group, df_O_Group, df_W_Group, df_E_Group, df_R_Group, df_NCD_UNDER70]:
print(f'{df.shape[0]} rows and {df.shape[1]} columns')
4428 rows and 23 columns
1560 rows and 23 columns
1560 rows and 23 columns
1560 rows and 23 columns
1560 rows and 23 columns
1560 rows and 23 columns
1365 rows and 23 columns
11640 rows and 23 columns
As df_R_Group has less rows than the other groups, after joining I’ll need to fill NaN values with zeros and make the notation is when displaying this data. Fortunately, the score goes from 1 to 6, then use 0 is not mislading.
Prepocessing
Keep just the columns of interest and renamed to better communication
Countries
# Countries dataframe
countries.head(2)
| Code | Title | Dimension | ParentDimension | ParentCode | ParentTitle | |
|---|---|---|---|---|---|---|
| 0 | ABW | Aruba | COUNTRY | REGION | AMR | Americas |
| 1 | AFG | Afghanistan | COUNTRY | REGION | EMR | Eastern Mediterranean |
# Rename the columns
countries.rename(columns = {'Code': 'Country Code', 'Title': 'Country', 'ParentTitle': 'WHO Region'}, inplace = True)
# Select specific columns using the .loc indexer
final_countries = countries.loc[:, ['Country Code', 'Country', 'WHO Region']]
# Sanity check
final_countries.head(3)
| Country Code | Country | WHO Region | |
|---|---|---|---|
| 0 | ABW | Aruba | Americas |
| 1 | AFG | Afghanistan | Eastern Mediterranean |
| 2 | AGO | Angola | Africa |
Estimate Average Smoking Prevalence
# Estimate of current tobacco use prevalence (%) dataframe
df_M_Est_smk_curr_std.head(1)
| Id | IndicatorCode | SpatialDimType | SpatialDim | TimeDimType | TimeDim | Dim1Type | Dim1 | Dim2Type | Dim2 | ... | DataSourceDim | Value | NumericValue | Low | High | Comments | Date | TimeDimensionValue | TimeDimensionBegin | TimeDimensionEnd | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 27794728 | M_Est_smk_curr_std | COUNTRY | DZA | YEAR | 2020 | SEX | BTSX | None | None | ... | None | 15.2 | 15.2 | None | None | Projected from surveys completed prior to 2020. | 2022-01-17T09:30:38.763+01:00 | 2020 | 2020-01-01T00:00:00+01:00 | 2020-12-31T00:00:00+01:00 |
1 rows × 23 columns
# Rename the columns
df_M_Est_smk_curr_std.rename(columns = {'SpatialDim': 'Country Code', 'ParentTitle': 'WHO Region', 'TimeDim' : 'Year', 'Dim1': 'Sex', 'Value': 'Prevalence'}, inplace = True)
# Select specific columns using the .loc indexer
final_prevalence = df_M_Est_smk_curr_std.loc[:, ['Country Code', 'Year', 'Sex', 'Prevalence']]
# Sanity check
final_prevalence.head(3)
| Country Code | Year | Sex | Prevalence | |
|---|---|---|---|---|
| 0 | DZA | 2020 | BTSX | 15.2 |
| 1 | BEN | 2020 | BTSX | 4.7 |
| 2 | BWA | 2020 | BTSX | 15 |
MPOWER indicators
# Rename the columns
df_M_Group.rename(columns = {'SpatialDim': 'Country Code', 'ParentTitle': 'WHO Region', 'TimeDim':'Year', 'Value': 'M_score'}, inplace = True)
df_P_Group.rename(columns = {'SpatialDim': 'Country Code', 'ParentTitle': 'WHO Region', 'TimeDim':'Year', 'Value': 'P_score'}, inplace = True)
df_O_Group.rename(columns = {'SpatialDim': 'Country Code', 'ParentTitle': 'WHO Region', 'TimeDim':'Year', 'Value': 'O_score'}, inplace = True)
df_W_Group.rename(columns = {'SpatialDim': 'Country Code', 'ParentTitle': 'WHO Region', 'TimeDim':'Year', 'Value': 'W_score'}, inplace = True)
df_E_Group.rename(columns = {'SpatialDim': 'Country Code', 'ParentTitle': 'WHO Region', 'TimeDim':'Year', 'Value': 'E_score'}, inplace = True)
df_R_Group.rename(columns = {'SpatialDim': 'Country Code', 'ParentTitle': 'WHO Region', 'TimeDim':'Year', 'Value': 'R_score'}, inplace = True)
# Select specific columns using the .loc indexer
final_M = df_M_Group.loc[:, ['Country Code', 'Year', 'M_score']]
final_P = df_P_Group.loc[:, ['Country Code', 'Year', 'P_score']]
final_O = df_O_Group.loc[:, ['Country Code', 'Year', 'O_score']]
final_W = df_W_Group.loc[:, ['Country Code', 'Year', 'W_score']]
final_E = df_E_Group.loc[:, ['Country Code', 'Year', 'E_score']]
final_R = df_R_Group.loc[:, ['Country Code', 'Year', 'R_score']]
Premature deaths due to noncommunicable diseases (NCD) as a proportion of all NCD deaths
# NDC: Noncommunicable diseases
df_NCD_UNDER70.head(1)
| Id | IndicatorCode | SpatialDimType | SpatialDim | TimeDimType | TimeDim | Dim1Type | Dim1 | Dim2Type | Dim2 | ... | DataSourceDim | Value | NumericValue | Low | High | Comments | Date | TimeDimensionValue | TimeDimensionBegin | TimeDimensionEnd | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 25578824 | NCD_UNDER70 | COUNTRY | AFG | YEAR | 2000 | SEX | MLE | GHECAUSES | GHE060 | ... | None | 72.8 [55.8-83.5] | 72.78874 | 55.84165 | 83.46061 | None | 2021-03-10T15:22:19.27+01:00 | 2000 | 2000-01-01T00:00:00+01:00 | 2000-12-31T00:00:00+01:00 |
1 rows × 23 columns
# Rename the columns
df_NCD_UNDER70.rename(columns = {'SpatialDim': 'Country Code','TimeDim':'Year', 'Dim1':'Sex', 'NumericValue':'PrematureDeaths/NCD'}, inplace = True)
# Select specific columns using the .loc indexer
final_premature_deaths = df_NCD_UNDER70.loc[:, ['Country Code', 'Year', 'Sex', 'PrematureDeaths/NCD']]
# Sanity check
final_premature_deaths.head(3)
| Country Code | Year | Sex | PrematureDeaths/NCD | |
|---|---|---|---|---|
| 0 | AFG | 2000 | MLE | 72.78874 |
| 1 | AFG | 2001 | MLE | 72.56174 |
| 2 | AFG | 2002 | MLE | 72.96648 |
Merging dataframes
Now concatenate the dataframes, it would be great to build just 1 dataframe. However, having some columns with more and less data, concatenating all of them would get NaN values and not option to set correct datatypes.
# Merging the DataFrames: final_countries, final_prevalence, final_MPOWER, and final_premature_deaths.
final_prevalence = final_countries.merge(final_prevalence, on='Country Code', how='inner')
final_premature_deaths = final_countries.merge(final_premature_deaths, on=['Country Code'], how='inner')
final_MPOWER = final_countries.merge(final_M, on=['Country Code'], how='inner')
final_MPOWER = final_MPOWER.merge(final_P, on=['Country Code', 'Year'], how='inner')
final_MPOWER = final_MPOWER.merge(final_O, on=['Country Code', 'Year'], how='inner')
final_MPOWER = final_MPOWER.merge(final_W, on=['Country Code', 'Year'], how='inner')
final_MPOWER = final_MPOWER.merge(final_E, on=['Country Code', 'Year'], how='inner')
final_MPOWER = final_MPOWER.merge(final_R, on=['Country Code', 'Year'], how='outer')
final_MPOWER
| Country Code | Country | WHO Region | Year | M_score | P_score | O_score | W_score | E_score | R_score | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | AFG | Afghanistan | Eastern Mediterranean | 2007 | 1 | 3 | 3 | 2 | 4 | NaN |
| 1 | AFG | Afghanistan | Eastern Mediterranean | 2008 | 1 | 3 | 3 | 2 | 4 | 2 |
| 2 | AFG | Afghanistan | Eastern Mediterranean | 2010 | 1 | 3 | 3 | 2 | 4 | 2 |
| 3 | AFG | Afghanistan | Eastern Mediterranean | 2012 | 1 | 3 | 3 | 2 | 4 | 2 |
| 4 | AFG | Afghanistan | Eastern Mediterranean | 2014 | 2 | 3 | 3 | 2 | 4 | 3 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1555 | ZWE | Zimbabwe | Africa | 2012 | 1 | 3 | 3 | 2 | 2 | 3 |
| 1556 | ZWE | Zimbabwe | Africa | 2014 | 2 | 3 | 2 | 2 | 2 | 3 |
| 1557 | ZWE | Zimbabwe | Africa | 2016 | 2 | 3 | 4 | 2 | 2 | 3 |
| 1558 | ZWE | Zimbabwe | Africa | 2018 | 2 | 3 | 4 | 2 | 2 | 3 |
| 1559 | ZWE | Zimbabwe | Africa | 2020 | 1 | 3 | 4 | 2 | 2 | 3 |
1560 rows × 10 columns
# Check values in R_score:
final_MPOWER['R_score'].unique()
array([nan, '2', '3', '1', '4', '5', 'Not applicable'], dtype=object)
# Replace 'Not applicable' with '0' in 'R_score'
final_MPOWER['R_score'] = final_MPOWER['R_score'].replace('Not applicable', '0')
# Fill NaN values with '0'
final_MPOWER['R_score'] = final_MPOWER['R_score'].fillna('0')
# Sanity check
final_MPOWER['R_score'].unique()
array(['0', '2', '3', '1', '4', '5'], dtype=object)
final_prevalence
| Country Code | Country | WHO Region | Year | Sex | Prevalence | |
|---|---|---|---|---|---|---|
| 0 | AFG | Afghanistan | Eastern Mediterranean | 2020 | BTSX | 9.5 |
| 1 | AFG | Afghanistan | Eastern Mediterranean | 2020 | MLE | 16.7 |
| 2 | AFG | Afghanistan | Eastern Mediterranean | 2020 | FMLE | 2.2 |
| 3 | AFG | Afghanistan | Eastern Mediterranean | 2005 | BTSX | 17.1 |
| 4 | AFG | Afghanistan | Eastern Mediterranean | 2005 | MLE | 27.5 |
| ... | ... | ... | ... | ... | ... | ... |
| 4423 | ZWE | Zimbabwe | Africa | 2025 | FMLE | 0.6 |
| 4424 | ZWE | Zimbabwe | Africa | 2000 | BTSX | 19.5 |
| 4425 | ZWE | Zimbabwe | Africa | 2000 | MLE | 36.1 |
| 4426 | ZWE | Zimbabwe | Africa | 2000 | FMLE | 2.9 |
| 4427 | ZWE | Zimbabwe | Africa | 2015 | FMLE | 1.2 |
4428 rows × 6 columns
# Check datatypes
final_prevalence.info()
final_premature_deaths.info()
final_MPOWER.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 4428 entries, 0 to 4427
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Country Code 4428 non-null object
1 Country 4428 non-null object
2 WHO Region 4428 non-null object
3 Year 4428 non-null int64
4 Sex 4428 non-null object
5 Prevalence 4428 non-null object
dtypes: int64(1), object(5)
memory usage: 242.2+ KB
<class 'pandas.core.frame.DataFrame'>
Int64Index: 10980 entries, 0 to 10979
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Country Code 10980 non-null object
1 Country 10980 non-null object
2 WHO Region 10980 non-null object
3 Year 10980 non-null int64
4 Sex 10980 non-null object
5 PrematureDeaths/NCD 10980 non-null float64
dtypes: float64(1), int64(1), object(4)
memory usage: 600.5+ KB
<class 'pandas.core.frame.DataFrame'>
Int64Index: 1560 entries, 0 to 1559
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Country Code 1560 non-null object
1 Country 1560 non-null object
2 WHO Region 1560 non-null object
3 Year 1560 non-null int64
4 M_score 1560 non-null object
5 P_score 1560 non-null object
6 O_score 1560 non-null object
7 W_score 1560 non-null object
8 E_score 1560 non-null object
9 R_score 1560 non-null object
dtypes: int64(1), object(9)
memory usage: 134.1+ KB
# Fix datatypes in M_POWER dataframe
final_prevalence['Prevalence'] = final_prevalence['Prevalence'].astype(float)
final_MPOWER['M_score'] = final_MPOWER['M_score'].astype(int)
final_MPOWER['P_score'] = final_MPOWER['P_score'].astype(int)
final_MPOWER['O_score'] = final_MPOWER['O_score'].astype(int)
final_MPOWER['W_score'] = final_MPOWER['W_score'].astype(int)
final_MPOWER['E_score'] = final_MPOWER['E_score'].astype(int)
final_MPOWER['R_score'] = final_MPOWER['R_score'].astype(int)
# Sanity check
final_prevalence.info()
final_MPOWER.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 4428 entries, 0 to 4427
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Country Code 4428 non-null object
1 Country 4428 non-null object
2 WHO Region 4428 non-null object
3 Year 4428 non-null int64
4 Sex 4428 non-null object
5 Prevalence 4428 non-null float64
dtypes: float64(1), int64(1), object(4)
memory usage: 242.2+ KB
<class 'pandas.core.frame.DataFrame'>
Int64Index: 1560 entries, 0 to 1559
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Country Code 1560 non-null object
1 Country 1560 non-null object
2 WHO Region 1560 non-null object
3 Year 1560 non-null int64
4 M_score 1560 non-null int64
5 P_score 1560 non-null int64
6 O_score 1560 non-null int64
7 W_score 1560 non-null int64
8 E_score 1560 non-null int64
9 R_score 1560 non-null int64
dtypes: int64(7), object(3)
memory usage: 134.1+ KB
Great! Now is easier to deal with the data and build the graphs to the dashboard.
Visualizations
Note: Values from 2000 to 2018 are estimated based on national surveys. From 2020 to 2025 the values are projections.
This section includes some of the visualizations, if you want to see all of them you can explore the GitHub repository.
Estimate Average Smoking Prevalence by Sex over the time
# Group by Sex and Year and calculate the mean of Prevalence
average_numeric_value_over_years = final_prevalence.groupby(['Sex', 'Year'])['Prevalence'].mean().reset_index()
pivot_table = pd.pivot_table(
average_numeric_value_over_years,
values='Prevalence',
index='Year',
columns='Sex',
aggfunc='mean'
)
pivot_table.reset_index(inplace=True)
fig = px.line(
pivot_table,
x='Year',
y=pivot_table.columns[1:],
title='Estimate Average Smoking Prevalence by Sex over the time'
)
fig.update_layout(
xaxis_title='Year',
yaxis_title='Prevalence (%)',
)
fig.show()
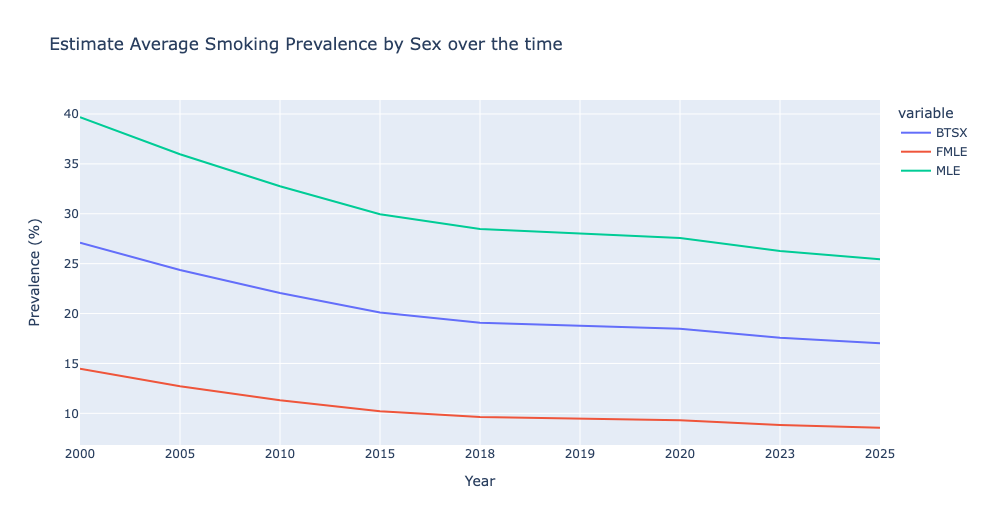
- Estimate prevalence in males is ~3 times bigger than in females group.
- The trend in the prevalence by sex groups is to decline. The prevalence projected by 2025 is ~15% lesss than the estimation in 2020. Females are expected to have a reduction of 6% in 2025.
Estimate Average Smoking Prevalence by Country over the years
# Sort the dataframe by Year in ascending order
aggregated_data = final_prevalence.sort_values('Year')
fig = px.choropleth(
aggregated_data,
locations='Country',
locationmode='country names',
color='Prevalence',
hover_name='Country',
animation_frame='Year',
color_continuous_scale='Viridis_r',
title='Estimate Average Smoking Prevalence by Country over the years',
)
fig.update_geos(
showcoastlines=True,
coastlinecolor='RebeccaPurple',
showland=True,
landcolor='LightGrey',
showocean=True,
oceancolor='LightBlue',
projection_type='natural earth',
)
fig.update_layout(
geo=dict(showframe=False, showcoastlines=False),
coloraxis_colorbar=dict(title='Prevalence (%)'),
)
fig.show()
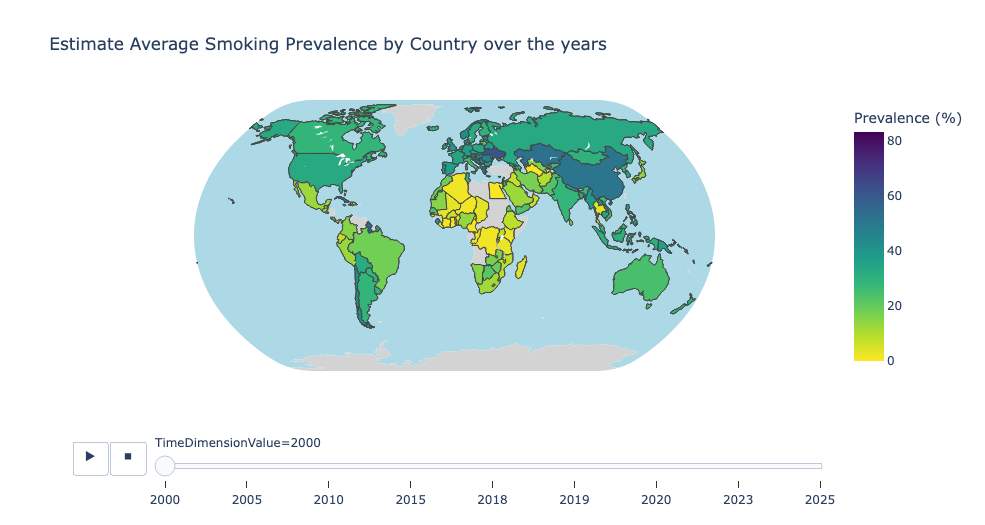
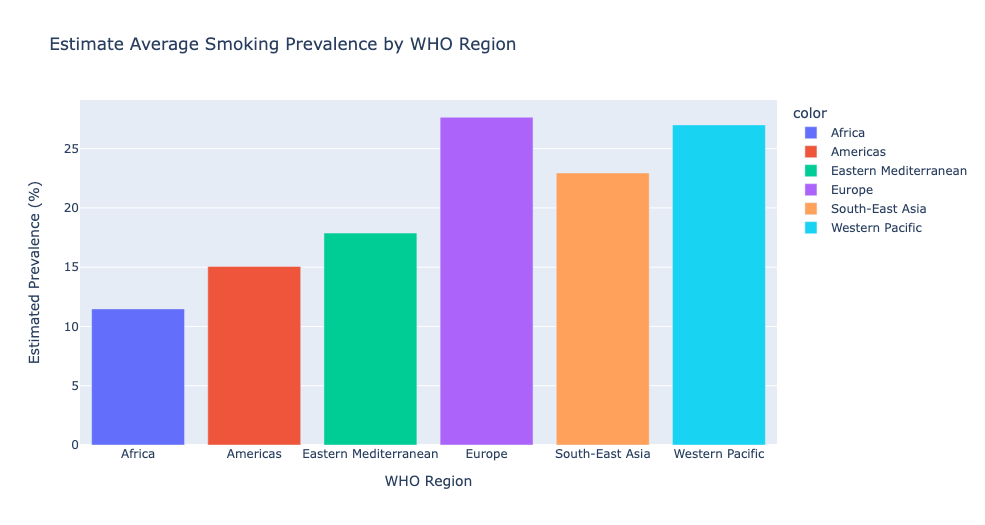
Estimate Average Smoking Prevalence by WHO Region
# Group by WHO Region and calculate the mean of Estimate Average Smoking Prevalence
average_numeric_value = final_prevalence.groupby('WHO Region')['Prevalence'].mean()
# Estimate Average Smoking Prevalence by WHO Region
fig = px.bar(
x=average_numeric_value.index,
y=average_numeric_value.values,
color=average_numeric_value.index,
labels={'x': 'WHO Region', 'y': 'Estimated Prevalence'},
title='Estimate Average Smoking Prevalence by WHO Region',
)
fig.update_layout(
xaxis_title='WHO Region',
yaxis_title='Estimated Prevalence (%)',
)
fig.show()
Get country MPOWER data function
Writting a function to get the indicator value for each country:
def get_country_data(final_MPOWER, country):
country_data = final_MPOWER[final_MPOWER['Country'] == country]
if country_data.empty:
return f'wrong spelling or "No data available for {country}'
specific_data = country_data[['Country', 'Year', 'M_score', 'P_score', 'O_score', 'W_score', 'E_score', 'R_score' ]]
return specific_data
result = get_country_data(final_MPOWER, 'Canada')
result
| Country | Year | M_score | P_score | O_score | W_score | E_score | R_score | |
|---|---|---|---|---|---|---|---|---|
| 232 | Canada | 2007 | 4 | 5 | 3 | 4 | 2 | 0 |
| 233 | Canada | 2008 | 4 | 5 | 5 | 4 | 2 | 4 |
| 234 | Canada | 2010 | 4 | 5 | 5 | 4 | 4 | 4 |
| 235 | Canada | 2012 | 4 | 5 | 5 | 5 | 4 | 4 |
| 236 | Canada | 2014 | 4 | 5 | 5 | 5 | 4 | 4 |
| 237 | Canada | 2016 | 4 | 5 | 5 | 5 | 4 | 4 |
| 238 | Canada | 2018 | 4 | 5 | 5 | 5 | 4 | 4 |
| 239 | Canada | 2020 | 4 | 5 | 5 | 5 | 4 | 4 |
Relation Smoking Prevalence vs Premature Deaths by Noncommunicable Diseases (NDC) (proportion among all NCD)
# Merging data to the scatterplot
scatter_data = pd.merge(final_prevalence, final_premature_deaths, on=['Country Code', 'Country', 'WHO Region', 'Year', 'Sex'])
scatter_data
| Country Code | Country | WHO Region | Year | Sex | Prevalence | PrematureDeaths/NCD | |
|---|---|---|---|---|---|---|---|
| 0 | AFG | Afghanistan | Eastern Mediterranean | 2005 | BTSX | 17.1 | 71.98372 |
| 1 | AFG | Afghanistan | Eastern Mediterranean | 2005 | MLE | 27.5 | 74.25545 |
| 2 | AFG | Afghanistan | Eastern Mediterranean | 2005 | FMLE | 6.7 | 69.67499 |
| 3 | AFG | Afghanistan | Eastern Mediterranean | 2010 | BTSX | 14.0 | 70.07750 |
| 4 | AFG | Afghanistan | Eastern Mediterranean | 2010 | MLE | 23.4 | 72.69907 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 2839 | ZWE | Zimbabwe | Africa | 2019 | FMLE | 0.9 | 56.25971 |
| 2840 | ZWE | Zimbabwe | Africa | 2000 | BTSX | 19.5 | 56.86102 |
| 2841 | ZWE | Zimbabwe | Africa | 2000 | MLE | 36.1 | 59.35542 |
| 2842 | ZWE | Zimbabwe | Africa | 2000 | FMLE | 2.9 | 54.56320 |
| 2843 | ZWE | Zimbabwe | Africa | 2015 | FMLE | 1.2 | 58.01034 |
2844 rows × 7 columns
Pearson Correlation Coefficient (Pearson’s r)
from scipy import stats
prevalence = scatter_data['Prevalence']
deaths = scatter_data['PrematureDeaths/NCD']
pearson_corr = stats.pearsonr(prevalence, deaths)
pearson_corr
PearsonRResult(statistic=-0.09868028979854496, pvalue=1.3413969700439424e-07)
The correlation between these two variables suggests a weak negative correlation statistically significant.
# Scatterplot of Smoking Prevalence vs Premature Deaths
fig = px.scatter(
scatter_data,
x='Prevalence',
y='PrematureDeaths/NCD',
title='Relation Smoking Prevalence vs Premature Deaths by NCD (proportion among all NCD)',
labels={'Prevalence': 'Smoking Prevalence', 'PrematureDeaths/NCD': 'Premature Deaths'},
animation_frame='Year',
color = 'WHO Region',
width=850,
height=850
)
# Set fixed axis ranges
fig.update_xaxes(range=[0, 100])
fig.update_yaxes(range=[0, 100])
# Show the plot
fig.show()
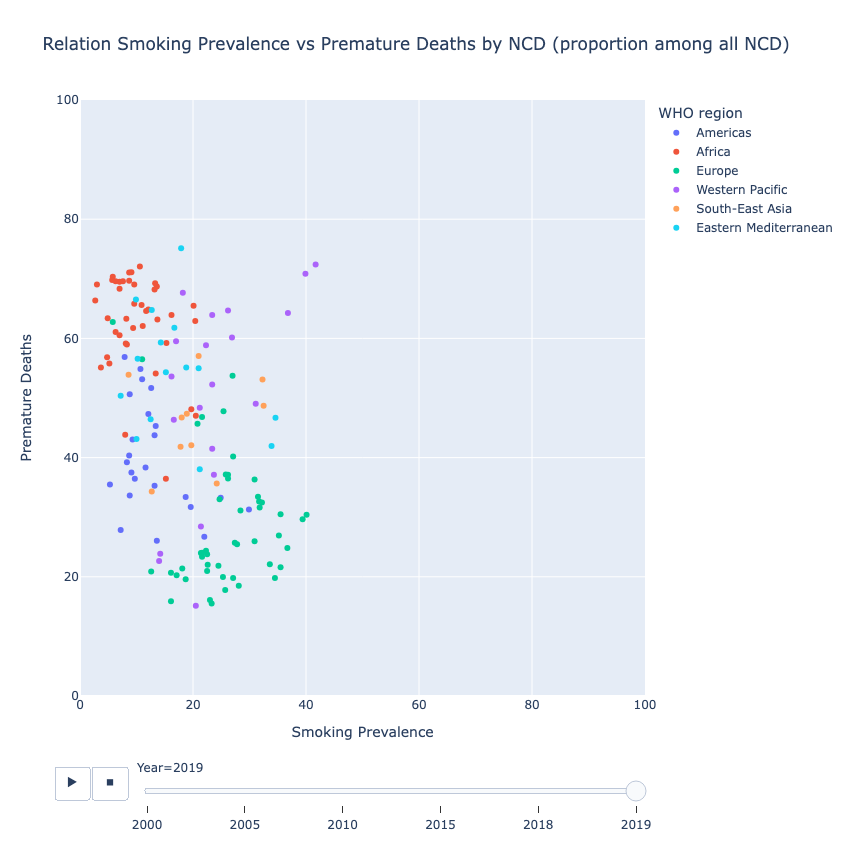
Design the Application/Dashboard
Now let’s write the csv files to work with them in the python script where the application is going to be defined.
# final_prevalence.to_csv('final_prevalence.csv')
# final_premature_deaths.to_csv('final_premature_deaths.csv')
# final_MPOWER.to_csv('final_MPOWER.csv')
# scatter_data.to_csv('scatter_data.csv')
I’m going to create a Python script for building the dashboard using the Dash framework.
I chose Dash for building the dashboard due to its seamless integration with Python and its ability to create interactive web applications for data visualization with relative ease. Dash’s Python-centric approach means I can leverage my existing Python programming skills without having to learn new languages. Its declarative syntax for defining the layout and interactions simplifies the development process. Additionally, Dash offers a wide range of pre-built components for creating charts, graphs, and interactive elements, reducing the need for extensive custom coding. The framework’s active community and extensive documentation provide a wealth of resources to troubleshoot issues and learn best practices. Overall, Dash’s combination of simplicity, versatility, and compatibility with Python makes it a powerful choice for rapidly developing engaging and interactive dashboards.
Overal Process View
First, I’ll make sure I have Dash installed. Then, I’ll set up my Python script, which I’ll name application.py, and import the necessary libraries. With the app initialized, I’ll define the layout using HTML and Dash components, structuring the interface the way I want it. To make the dashboard interactive, I’ll use callbacks that respond to user inputs and update the displayed content accordingly. Once everything is set up, I’ll run the app command and access it through my web browser. This process will allow me to create a dynamic and interactive dashboard for data visualization and analysis.
Dashboard description
As I explained, after connecting with GHO API, the exploration derived in the building of the dashboard: Global Tobacco Insights: Visualizing Smoking Trends, Health Consequences, and Control Policies a dashboard that display important information about Smoking Prevalence by sex, countries and WHO regions, the relationship between smoking rates and premature deaths caused by non-communicable diseases (NCDs), and the efforts being undertaken by the countries to accomplish the MPOWER strategy.
The dashboard provides insights into the challenges and successes of addressing smoking on a global scale. It highlights regions striving for improved public health and emphasizes the commitment of the global community to tackle this critical issue. Nevertheless, this project also underscored the need for additional data to gain a more comprehensive understanding of the smoking landscape. This is particularly critical given that the most recent data available from the Global Health Observatory (GHO) for constructing this project dates back to 2020.
Personal comment: The subjective perception doesn’t align with reality either, as it assumes that I live in a country with a low smoking prevalence. However, my personal experience contradicts this assumption, as I regularly encounter situations where I inhale secondhand smoke from individuals on the streets, which wouldn’t be expected in a country with such low prevalence and really good Scores in Protecting strategy. This situation prompts me to consider the possibility that we might not be accurately measuring the actual consumption rates and policies accomplishment. Absolutely, there is a clear need for more data and dedicated efforts to address this public health issue.
Deploy the Application/Dashboard
I’ll start by pushing my application.py script and all the project files to this repository. To ensure a clean and reproducible environment, I’ll include a requirements.txt file listing all the dependencies, including Dash and other libraries used in the project.
Next, I’ll utilize EC2 AWS instance to deploy the dashboard. I’ll create the instance and configure it to host the app.
Steps to deploy the dash application using EC2 AWS instance
- Log in on AWS.
- Select in
Services:EC2 - Launch a new instance and choose an Amazon Machine Image on your convenience, I chose a AMI and instance type with free tier because its capacity was enough for deploying my project.
- If you have not connected before with AWS you will need to create the Key Pair.
- When instance state be
Runninggo toinstance settings->Securityand change the include a newport. My case the new port was 8050 as that was the port dash used to deploy the app. - Connect to your instance using SSH protocol.
- Install the require libraries in your instance.
- Move your the files you require to run your app to your instance, I cloned the repository in my instance.
- I had to include Flask in my .py script and modify the code to access Dash app using the external IP address or domain name of the EC2 instance.
- Run the application in your instance.
- In Instance
Detailsyou will find thePublic IPv4 address, add:8050to that address to see the application running. - Install TMUX library and
Detachthe dash session to keep the app running even when you disconnect from the instance.
Application link
Sources
- Global Health Observatory
- GHO OData API
- World Health Organization - MPOWER measures
- Canada’s Tobacco Strategy
Acknowledgments
- Dash Python User Guide: Dash is the original low-code framework for rapidly building data apps in Python. Learn more here
- Dash Bootstrap Components: dash-bootstrap-components is a library of Bootstrap components for Plotly Dash, that makes it easier to build consistently styled apps with complex, responsive layouts. Learn more here
- Rahul Agarwal How to Deploy a Streamlit App using an Amazon Free ec2 instance?
